Tag Archives: editing
Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR–Cas9 ribonucleoprotein
Technology tamfitronics
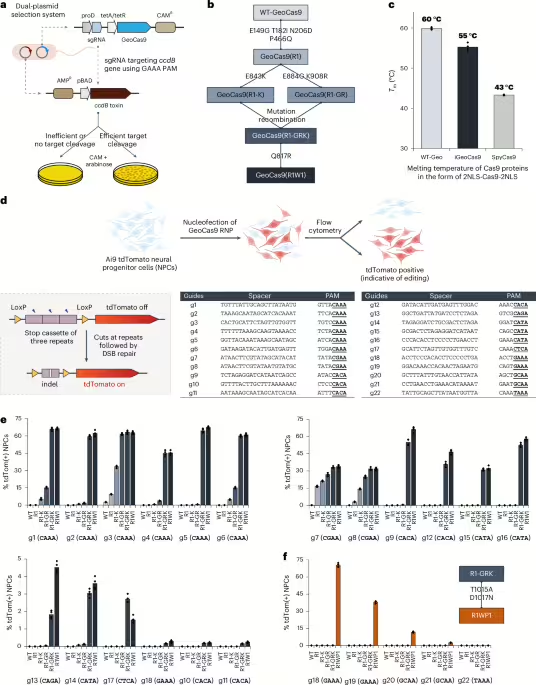
MainClustered regularly interspaced short palindromic repeats (CRISPR)–Cas9-based genome editing1,2,3 has the potential to provide wide-ranging treatments for genetic diseases4,5,6 if safe and effective methods for delivering CRISPR-based therapeutics can be developed7,8. Although viral delivery of CRISPR genome editors is the most widely used method for in vivo cell editing9,10,11, viral vectors can be immunogenic, carry the risk of vector genome integration and can induce off-target DNA damage because of continuous genome editor expression12. Alternative nonviral strategies for delivering CRISPR editors could address these limitations if issues of efficacy and toxicity can be overcome.Lipid nanoparticle (LNP)–mRNA complexes are nonvirally derived vehicles for in vivo delivery that have been remarkably successful at genome editing in...
Genome editing with DNA-dependent polymerases
Technology tamfitronics News & Views ...
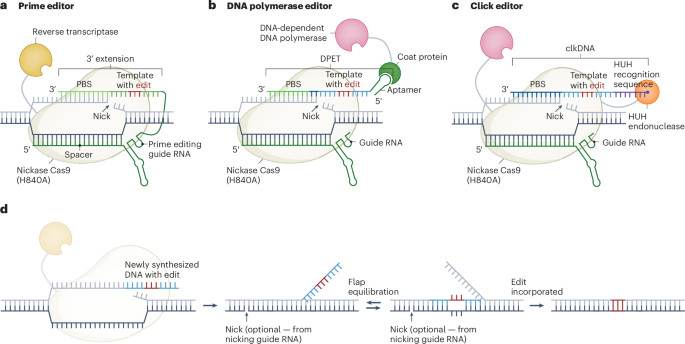
Click editing enables programmable genome writing using DNA polymerases and HUH endonucleases
Technology tamfitronics Doudna, J. A. The promise and challenge of therapeutic genome editing. Nature 578229–236 (2020).Article CAS PubMed PubMed Central Google Scholar Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38824–844 (2020).Article CAS PubMed Google Scholar ...
Precise in vivo RNA base editing with a wobble-enhanced circular CLUSTER guide RNA
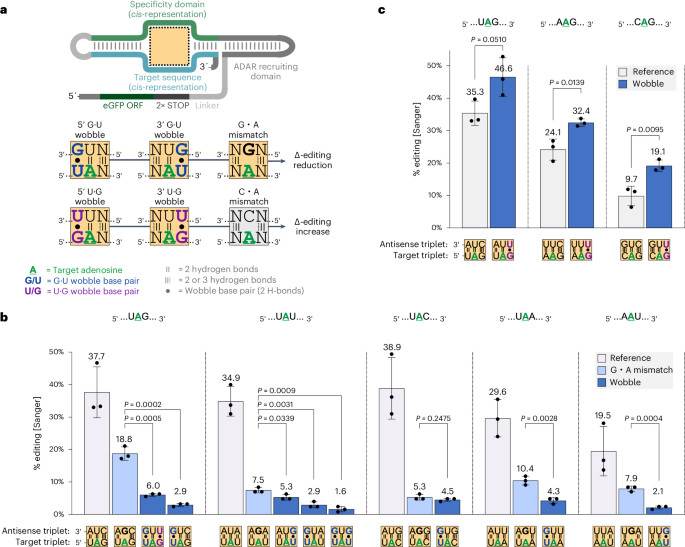
Technology tamfitronics MainSite-directed adenosine-to-inosine (A-to-I) RNA base editing is a very promising technology with a clear path for clinical application1,2. Hydrolytic deamination of A by enzymes of the ADAR family (adenosine deaminases acting on RNA) produces an inosine, which is biochemically interpreted as G in many cellular processes such as splicing or translation and, consequently, functionally substitutes A with G on the RNA level3,4. There are three catalytically active human ADAR proteins: constitutively and ubiquitously expressed ADAR1 p110, interferon-inducible ADAR1 p150 and ADAR2. In the past, ADAR deaminase domains have been engineered into various artificial editing approaches that enable the efficient and highly programmable editing of any given target A in the transcriptome by applying...